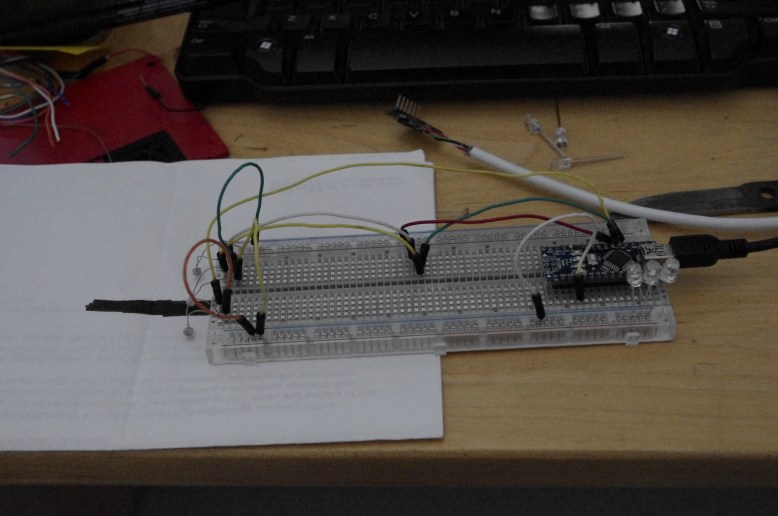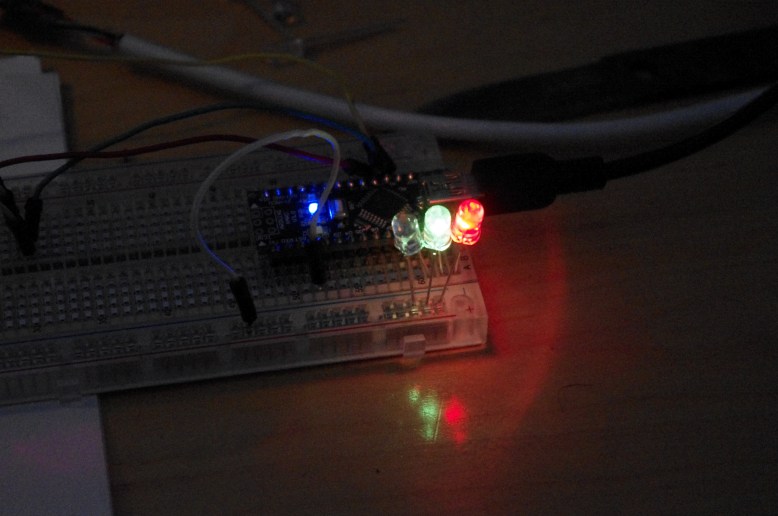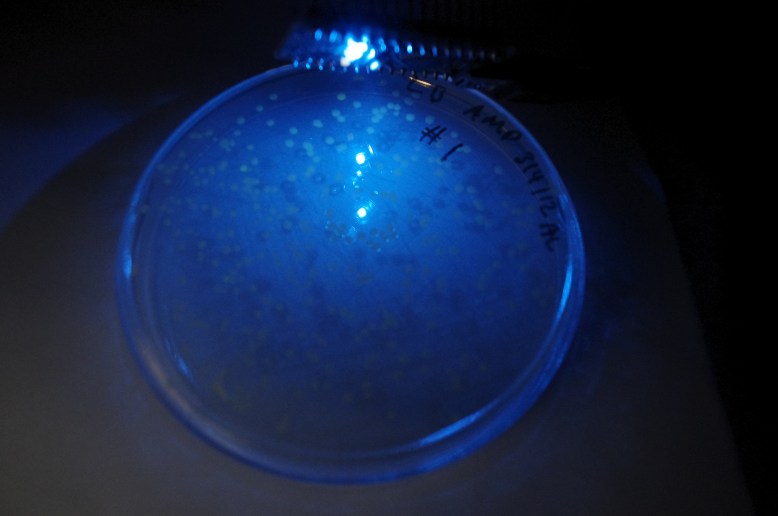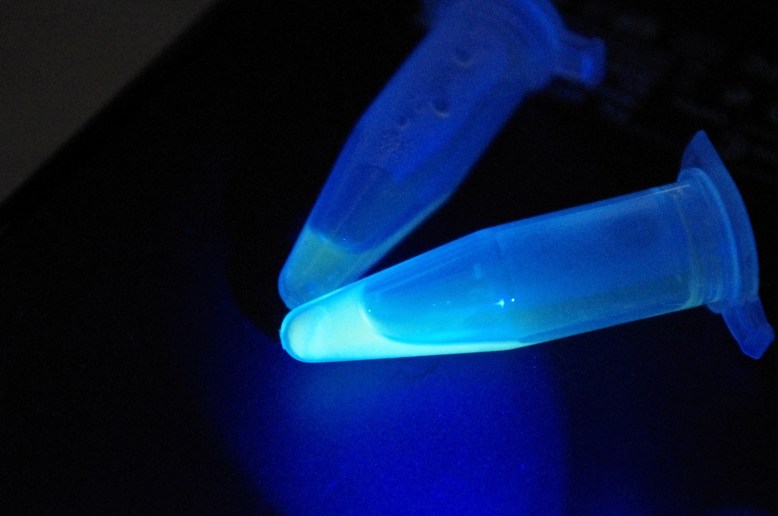
GFP! GFP has an excitation peak in the blue light spectrum. Unfortunately, the blue LED is comparatively bright. Gotta find a green pass filter!
This week was Week 2 of the GFP project! The extraction of the Green Fluorescent Protien (GFP) from the transformed host cells, using Hydrophobic Interaction Chromatography (HIC). We used a kit from Carolina because I have not ever done this before, and I didn’t really know what to buy to do this completely DIY style. Nonetheless, it was a success and a clear demonstration of the central dogma of biology (DNA–>RNA–>Protein), and a good way to add some value to our transformation. Now we have not only altered the bacteria, but made something pseudo-useful, but definitely cool, with our alteration.
At a high level, the way HIC works is by selectively binding the protein to a “bead” (more like a fine powder) resin. Once the cell has been lysed (the cell walls are broken up), the GFP-bearing lysate is mixed with a high salt binding solution. This gently and reversibly denatures the GFP so that the hydrophobic (water fearing) inside is exposed to the solution. This water fearing end acts like oil in water; it is attracted to other hydrophobic bits, so the bits all clump together. Since the beads have hydrophobic sites, they bind to lots and lots of proteins that are hydrophobic. At this stage, the liquid that contains non-binding proteins is removed from the matrix, so we remove it. Then a medium-salt wash buffer is added, which removes some of the hydrophobic proteins that re-fold in lower salt, and are therefore less “sticky” than the gfp. The extra liquid holding these proteins is then removed, and this is when the final low-salt TE buffer is added. The low salt buffer is also known as the elution buffer, because it elutes (washes out with a solvent) the GFP from the matrix by allowing the GFP to re-nature (fold back into its original configuration), and hide its hydrophobic sites. Then the liquid contains GFP! The process is described in more detail below.

Overnight cultures fluorescing! You want to use only a few mls for these, to increase the amount of oxygen-per-volume in each culture.
The first step is to grow overnight cultures of the transformants. To do this, aseptically pick off a colony from a plate of transformants that are (ideally) only a few days old, and use it to inoculate an LB+ampicillin liquid broth culture . The antibiotic keeps the selection pressure on the transformed cells, and doing it a few days after the transformation helps ensure that you do not pick satellite colonies or have e. coli that have dropped the plasmid. These are literally grown overnight, for about 12-16 hours in a shaker. It is important that the cells are exposed to oxygen, because it is critical to the development of the GFP. The cells should fluoresce brightly when blue light is shone on them. In the picture above, the middle and right tube are fluorescing green, but the rightmost tube is not.
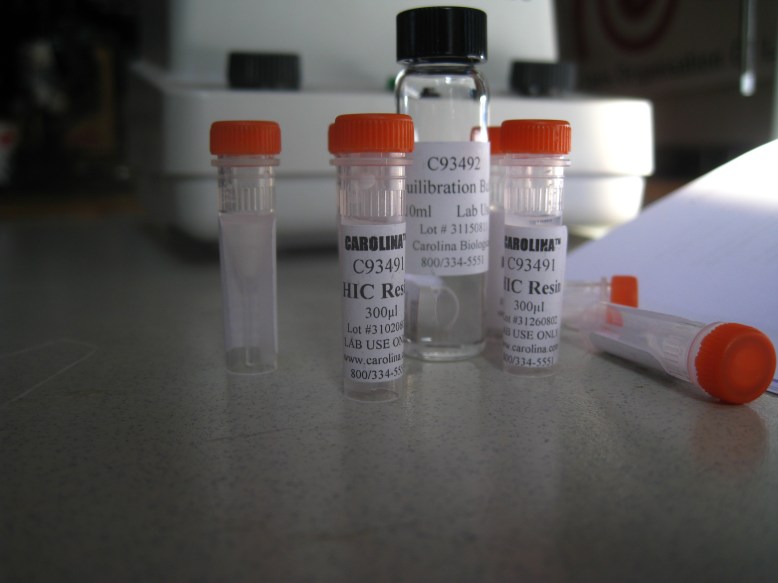
HIC resin and equilibration buffer
The next thing to do is to equilibrate the HIC resin with the equilibration buffer. This is just some salt so that when you add your bound GFP (cell lysate in high-salt solution), the water that the hydrophobic beads are suspended in does not bring the salinity down. To equilibrate, we added 300ul of HIC resin to 1mL of equilibration buffer, and centrifuged, removing the supernatant. The supernatant is the liquid remaining on top of the pellet (the mass of solids at the bottom of the microcentrifuge tube) once it has been centrifuged. Definitely remove the supernatant with a pipettor, because it is runny, so you need to keep the tube upright.
You can see the large green pellet at the bottom of the tube, after it has been centrifuged. The stuff on top is the supernatant.
The next steps you do twice. Add 1mL of overnight culture to a microcentrifuge tube, spin down for ~2 min at max RPM, being sure to balance the centrifuge. Then pour off the supernatant. Repeat for another 1mL

The big green thing at the bottom is a huge pellet of cells! Notice that the supernatant has been removed, and the cells are kind of stuck to one side of the tube. That is the side of the tube the was facing outwards.
At this point, we have a great big lump of green cells. Now we want to harvest the GFP, which is inside the cell membrane in the cytoplasm, with all the other cellular proteins. To get it out, we add a lysis buffer. I suspect that in this kit, it is SDS because it got bubbly/sudsy when we pipetted it.

Washboard technique!
The secret to lysing pelleted cells quickly is using the washboard technique. It works better (for me) than vortexing, and way better than flicking or pipetting up and down. What you do is grab the top of the eppendorf, and run it along the top of a tube rack, like you would run a stick against a washboard for musical effect. This subjects the tube to a lot of sudden shocks, and really blasts the pellet off the side of the tube. When this is done, the pellet and lysis buffer should not be clear; it should have a milky green consistency.

Lysate on ice. you can see the pale green lysate in the tubes.
Another factor in support of the lysis buffer being SDS and not lysosome enzymes is that the ice would slow the enzymatic reactions, while icing cells without any kind or cryoprotectant (like glycerol) is reputed to break up the cell membrane by forming ice crystals, which would further our goals in this stage.

Pellet again! This time the supernatant lysate is green, and contains the GFP we are trying to extract,
The lysate is taken off ice and centrifuged after 15 minutes, and 250 ul of supernatant is pipetted into a clean eppendorf. In this tube, we add the binding buffer, which gently denatures the GFP. It is called binding buffer because it allows the GFP to bind to the HIC resin, not because it binds to the GFP. It gets cloudy in this step, but still retains some greenness.

supernatant lysate in a clean tube. Note bubbles from (probably) SDS
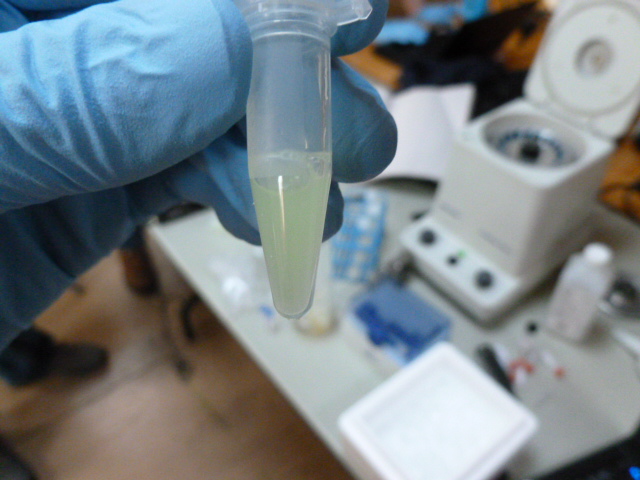
After the binding buffer has been added
The next step is to add the GFP bearing lysate in the binding buffer to the HIC resin. You will want to mix this thoroughly to ensure that the GFP binds to the resin.
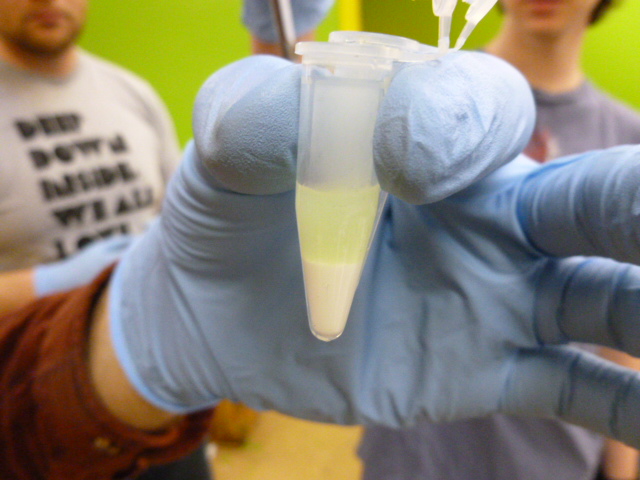
GFP in binding buffer sitting on the HIC resin
Once the beads and the lysate are mixed, you put the tubes into the centrifuge and pellet out the HIC resin, which is now bound to the GFP and many other proteins. As you can see in the picture below, the supernatant is not fluorescent, so it does not contain any significant amount of GFP.
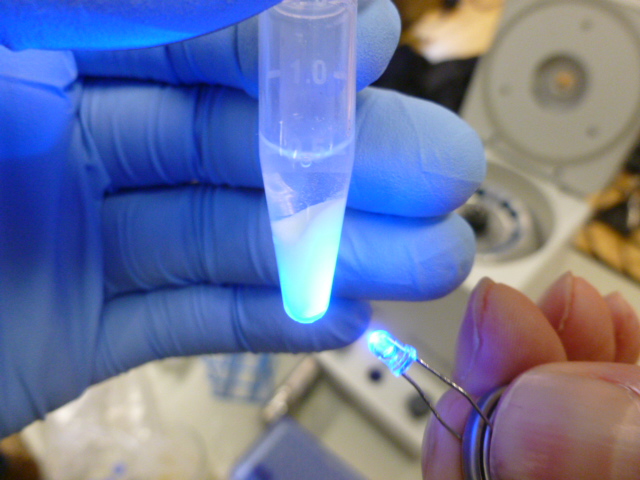
The resin glows now, but the supernatant does not!
Now the supernatant is pipetted off, and the wash buffer is added. The wash buffer is a less salty buffer than the binding buffer, so some of the proteins that are less sensitive to salt change configuration and “hide” their hydrophobic sites from the HIC resin, making them fall back into solution. This is centrifuged and the supernatant removed and discarded much like the binding buffer was.
The final step is to elute (to wash out with a solvent) the GFP from the resin with the TE (low salt) buffer. This makes the GFP fall off of the resin and into the solution. Then we pipetted the supernatant (now containing GFP) into a clean eppendorf. Here are some shots of the results:

left tube is the extract, right tube is the crude cell lysate

with blue light from my arduino!
Now next weekend the GFP project will be on hiatus, because I will be away for spring break. However, the sunday after that it will be back in full effect, probably with a plasmid DNA extraction adventure!